.: Core Research Activities in
Reconfigurable Computing Laboratory
|
7.
Many-Core Architecture Exploration for Scientific Applications
A)
iPlant Genotype-to-Phenotype (iPG2P) Grand Challenge Project ( Gregory
Streimer)
Elucidating
the relationship between plant genotypes and the resultant phenotypes
in complex (e.g., non-constant) environments is one of the foremost
challenges in plant biology. Plant phenotypes are determined by
often intricate interactions between genetic controls and environmental
contingencies. In a world where the environment is undergoing rapid,
anthropogenic change, predicting altered plant responses is central
to studies of plant adaptation, ecological genomics, crop improvement
activities (ranging from international agriculture to biofuels),
physiology (photosynthesis, stress, etc.), plant development, and
many many more. 
RCL
is participating in the NSF Funded
iPlant Collaborative by developing computationally intensive
algorithms to run on NVIDIA GPUs for relating genotype to phenotype
in complex environments.
High
throughput data analysis workflow in life sciences are typically
composed of iterative tasks that can potentially leverage the architectural
benefits afforded by GPGPUs to improve overall performance. The
compute bottlenecks in the workflow are interspersed and very often
are the rate limiting factors, it is important to consider the overall
execution environment and how GPGPU-based approaches can integrate
with the existing workflow. In collaboration with Prof. Steve Welch
(iPG2P) and designated working groups (Statistical Inference, Next
Gen), suitable candidate applications (algorithms) for exploration
will be identified. These identified application will involve solving
N equations with N unknowns. Matrix reduction and coefficient calculations
with large size vectors involve recurring sequences of instructions
(multiply-subtract, multiply-add, etc) within the loop level executions.
Traditional clusters can run these loops concurrently;
however, for largeN we expect GPGPUs to be a bettermatch to the
large number of fine-grain computations. The team at the University
of Arizona (Nirav Merchant, Arizona Research Lab, Prof. Ali Akoglu,
ECE), and (Prof. David Lowenthal, Computer Science) has been formed
to study the following aspects of GPGPUs:
- Evaluate
the effectiveness of Open CL and CUDA in developing GPGPU programs.
- Develop
architectural and application-level models to determine which
portions of a program to offload to the GPGPU.
- Explore
ways to restructure the program architecture in order to exploit
the unique memory hierarchy of the GPU and its processing power
with hundreds of processing cores.
- Demonstrate
the scalability of the developed algorithms to multiple GPUs.
- Quantify
the speed-up achieved with CUDA environment by performance comparison
against CPU based parallel system.
B)
Sequence Alignment (Student: Gregory Streimer and Yang song)
A common
problem posed in computational biology is the comparison of protein
sequences with unknown functionality to that of a set of known protein
 sequences to detect functional similarities. The most sensitive
algorithm for searching similarities is the Smith-Waterman algorithm,
however it is also the most time consuming. With an ever increasing
size in protein and DNA databases, searching them becomes more difficult
with exact algorithms such as Smith-Waterman. This research explores
reducing the computational time of the algorithm by utilizing multi-core
technologies. Currently we are examining the potential of Compute
Unified Device Architecture (CUDA) using the Nvidia C870 graphics
processing unit (GPU). This card has massively parallel computational
capabilities, which makes it ideal for high-performance scientific
computing. The C870 has sixteen multi-processors, each containing
eight streaming processors which individually operate at 1.35 GHz.
Each multi-processor is capable of launching up to 768 threads at
once, and each thread can run a Smith-Waterman alignment on a different
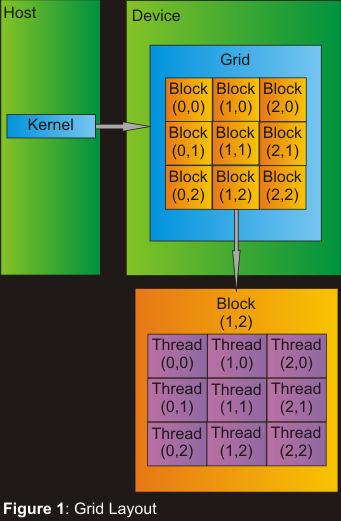
sequence pair. On the GPU, the threads are organized in blocks within
a grid of blocks which are launched from the kernel. This can be
seen in Figure 1.
sequences to detect functional similarities. The most sensitive
algorithm for searching similarities is the Smith-Waterman algorithm,
however it is also the most time consuming. With an ever increasing
size in protein and DNA databases, searching them becomes more difficult
with exact algorithms such as Smith-Waterman. This research explores
reducing the computational time of the algorithm by utilizing multi-core
technologies. Currently we are examining the potential of Compute
Unified Device Architecture (CUDA) using the Nvidia C870 graphics
processing unit (GPU). This card has massively parallel computational
capabilities, which makes it ideal for high-performance scientific
computing. The C870 has sixteen multi-processors, each containing
eight streaming processors which individually operate at 1.35 GHz.
Each multi-processor is capable of launching up to 768 threads at
once, and each thread can run a Smith-Waterman alignment on a different
sequence pair. On the GPU, the threads are organized in blocks within
a grid of blocks which are launched from the kernel. This can be
seen in Figure 1.
Current
limitations on the GPUs memory can constrain how much data can be
processed at any given time, so exploiting different memory resources
to their fullest potential for Smith-Waterman is also a part of
this research. Due to some of the memory restrictions, our implementation
utilizes the system's cached constant memory for storage of the
substitution matrices, as well as the user's query sequence. This
allows for faster access to these highly utilized values through
the use of an extremely efficient cost function. A major advantage
of our implementation over other GPU implementations is the fact
that we do not place restrictions on the length of database sequences,
so a database can truly be searched in its entirety. This work will
be benchmarked against another known GPU implementation, as well
as commonly used serial and multi-threaded CPU implementations.
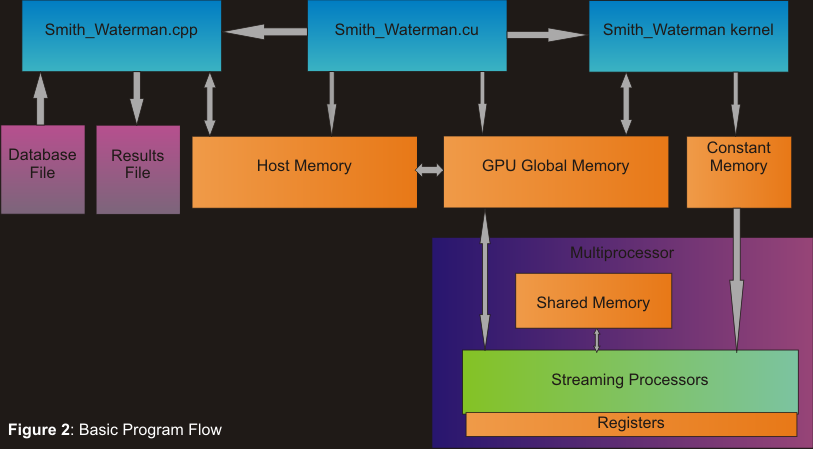
Figure 2 displays our basic program's flow
Publications:
- Yang
Song, Gregory M. Striemer and Ali Akoglu, "Performance Analysis
of IBM Cell Broadband Engine on Sequence Alignment", IEEE
NASA/ESA Conference on Adaptive Hardware and Systems (AHS 2009),
July 2009, San Francisco
- Greg
Streimer, Ali Akoglu, "Sequence Alignment with GPU: Performance
and Design Challenges", 23rd IEEE International Parallel
and Distributed Processing Symposium, May 25-29, 2009, Rome, Italy
This
study is posted on CUDA
Zone
Source
Code
fror Smith-Waterman Algorithm for use on an NVIDIA GPU using CUDA
(posted 6/16/2009)
|




