.: Core Research Activities in
Reconfigurable Computing Laboratory
|
5.
Many-Core Architecture Exploration for Scientific Applications
A) Accelerating the TCR Synthesis Process
Funded
by: NIH
Collaborator: Adam Buntzman (UA)
Student: Elnaz T. Yazdi
The TCR and BCR immune receptors have been developed into numerous therapies (e.g. monoclonal antibody therapy/mAb, adoptive T-cell therapies, and CAR T-cell therapies) that form the basis of the emerging fields of immunotherapy and immunodiagnostics.
The earliest clinical contributions in the field have already saved countless lives while garnering a Nobel Prize, yet the field is in its infancy and progress is hampered by a lack of understanding of the nature of which immune receptors are 1) created by the immune system, 2) bind to which antigenic targets, 3) become recruited into an immune response, and 4) retained in a memory anamnestic response.
The main hindrance to systematically developing new immunotherapies, new immunodiagnostics, and novel immunobiomarkers is the enormous magnitude (>10^18 unique species) of the repertoire of TCR and BCR species that can be made by the immune system. 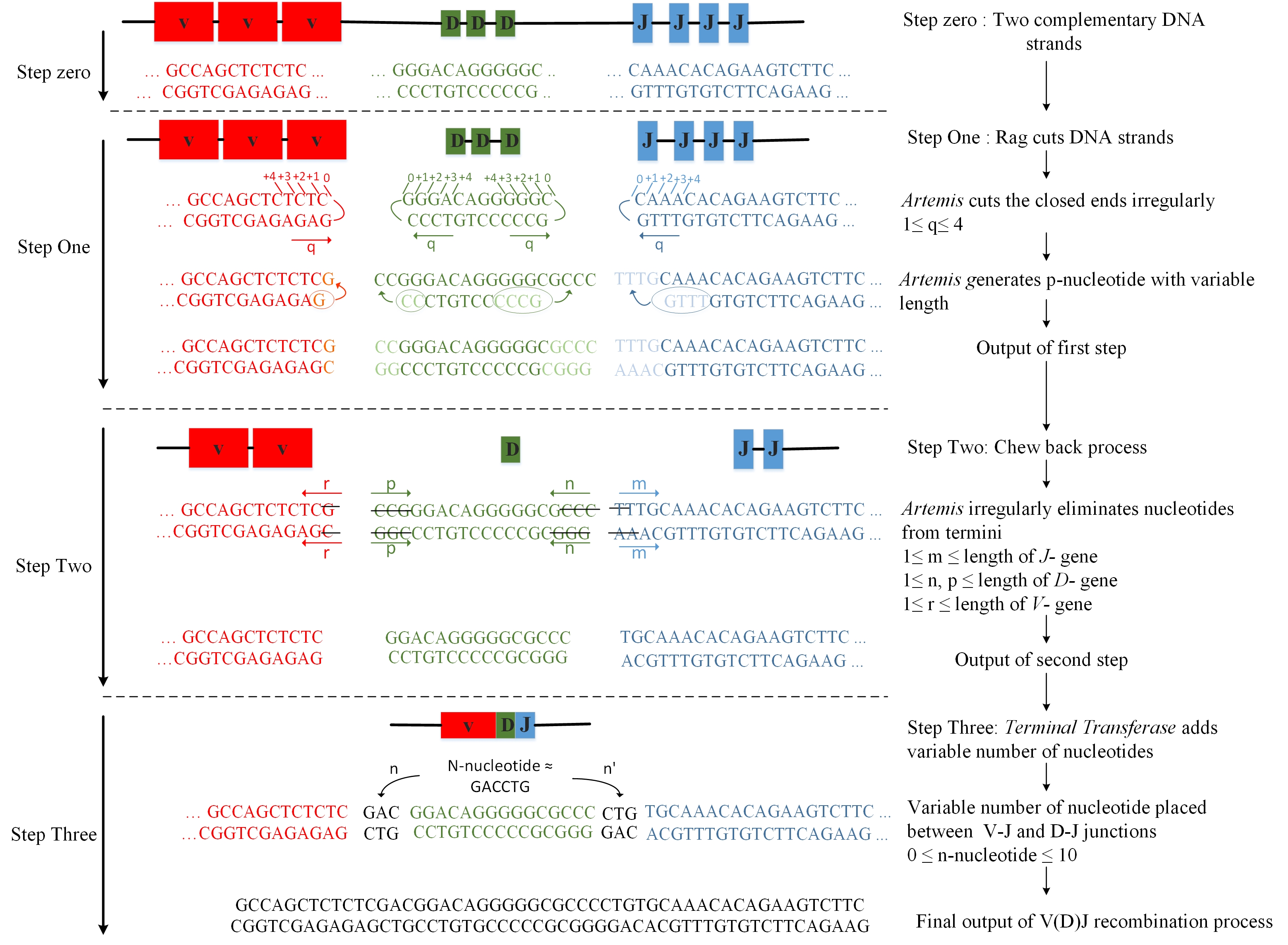
Computational tools with modeling and predictive power which can handle this massive scale are sorely needed. Therefore, we developed an GPU-based synthesis-on-the-fly approach to make the complete landscape of immune receptors available to be screened and modeled during in silico synthesis.
The most exhaustive study on TCR synthesis to this date models more than 10^14 TCR chains by exploiting the data parallel nature of the recombination process and mapping the algorithm entirely on a single graphic processor unit (GPU) [1]. The source code is available HERE
The study by Striemer et al.[1], which successfully models the mouse TCR repertoire for the first time, shows that the time scale of the TCR synthesis can be reduced to 16 days on a single NVIDIA GTX 480 GPU from an estimated execution time of 52 weeks on a general purpose processor. This study models all the potential TCR repertoire of the mouse in which the number of recombination pathways exceeds 4*10^14. Our aim is to investigate ways to reduce the execution time and memory footprint of the recombination process using mouse data set as a reference so that we establish a basis for rapidly modeling systems that are more complex than the mouse. Towards this goal, compared to the state of the art GPU based implementation, we make the following contributions:
- Bit-wise implementation of the recombination process, which consists of fine-grained shift, concatenation, comparison, and counting over the binary domain input data set. We show that the execution time of the baseline implementation reduces by a factor of 2.1 through bit-wise implementation on a single NVIDIA Tesla P100 GPU. The source code is available HERE
- Muti-GPU implementation with even workload distribution across the GPUs. For the multi-GPU implementation we introduce a task generation function that generates a unique task for each thread and eliminates the communication between the GPU threads. We conduct scalability analysis and show that the execution time of the baseline implementation reduces by a factor of 9.4 through bit-wise and multi-GPU implementation for the eight GPU configuration. The source code is available HERE
- We show that bit-wise implementation reduces the execution time from 40.5 hours to 18.9 hours on a single GPU and to 4.3 hours on an 8-GPU configuration. The data set is available HERE
Publications:
- Gregory Striemer, Harsha Krovi, Ali Akoglu, Benjamin Vincent, Ben Hopson, Jeffrey Frelinger, and Adam Buntzman, “Overcoming the Limitations Posed by TCR-beta Repertoire Modeling through a GPU-Based In-Silico DNA Recombination Algorithm,” In Proceedings of the 2014 IEEE 28th International Parallel and Distributed Processing Symposium (IPDPS '14), May 19-23, 2014, Phoenix, AZ, pp. 231-240.
- Benjamin Vincent, Adam Buntzman, Benjamin Hopson, Chris McEwen, Lindsay Cowell, Ali Akoglu, Helen Zhang, Jeffrey Frelinger, “iWAS – A novel approach to analyzing Next Generation Sequence data for immunology,” Cellular Immunology, Volume 299, January 2016, Pages 6-13, ISSN 0008-8749, http://dx.doi.org/10.1016/j.cellimm.2015.10.012.
- Khaled
Benkrid, Ali Akoglu, Cheng Ling, Yang Son, Ying Liu, and Xiang
Tian, "High performance biological pairwise sequence alignment:
FPGA vs. GPU vs. Cell BE vs. GPP," International Journal
of Reconfigurable Computing, special issue on "High Performance
Reconfigurable Computing," accepted for publication, 2011
- Yang
Song,
Gregory M. Striemer and Ali Akoglu, "Performance Analysis
of IBM Cell Broadband Engine on Sequence Alignment", IEEE
NASA/ESA Conference on Adaptive Hardware and Systems (AHS 2009),
July 2009, San Francisco
- Greg
Streimer, Ali Akoglu, "Sequence Alignment with GPU: Performance
and Design Challenges", 23rd IEEE International Parallel
and Distributed Processing Symposium, May 25-29, 2009, Rome, Italy
Archived sequence alignment work along with the srouce code is available here here and here
B)
Cardiac Simulation on Multi-GPU Platform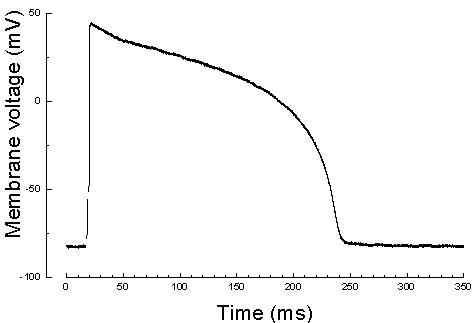 
Funded by: NSF (I/UCRC Cloud and Autonomic Computing)
Collaborator: Sarver Heart Center (Talal Moukabary, Steven Goldman, Elizabeth Juneman)
Student: Ehsan Esmaili
Chronic heart failure (CHF) occurs when the heart is damaged and unable to sufficiently pump blood throughout the body. CHF affects millions of Americans each year and it is the leading cause of hospitalization of the patients over the age of 65. In addition, cardiac arrhythmias and sudden cardiac death, specifically due to ventricular tachycardia (VT) and ventricular fibrillation (VF) in patients with CHF are among the most common causes of death in the industrialized world. Despite decades of research the relationship between CHF and VT/VF is poorly understood.
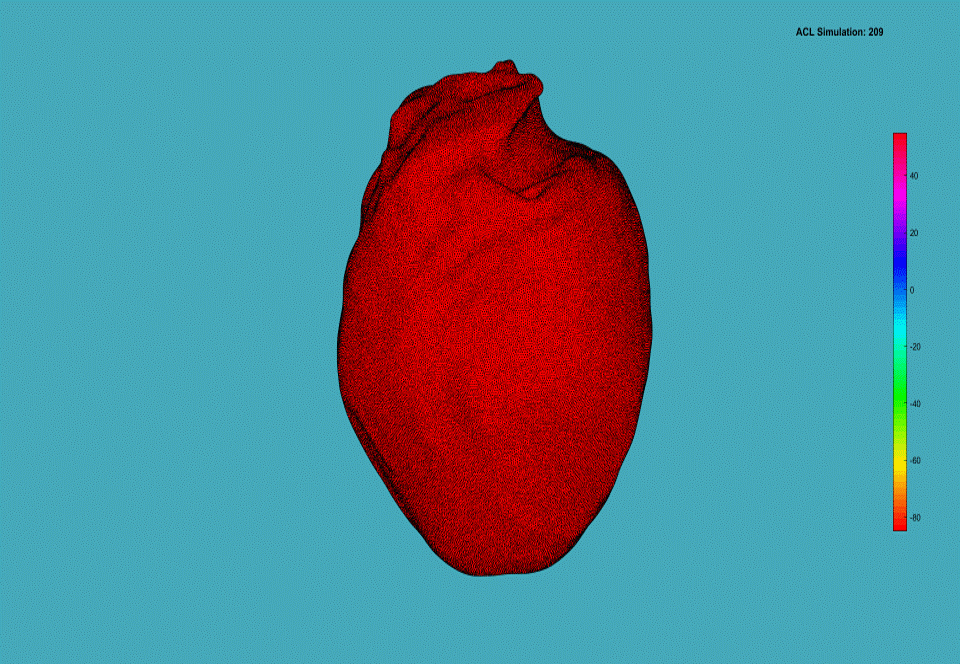
Computational models of the human cardiac cells provide detailed properties of human ventricular cells. The execution time for a realistic 3D heart simulation based on these models is a major barrier for physicians to study and understand the heart diseases, and evaluate hypotheses rapidly towards developing treatments. Computational models of the human cardiac cells exist and provide detailed properties of human ventricular cells, such as the major ionic currents, calcium transients, and action potential duration (APD) restitution, and important properties of wave propagation in human ventricular tissue, such as conduction velocity (CV) restitution (CVR). The complexity of detailed 3D models has led cardiac researchers to less accurate models, such as the monodomain model, that are computationally tractable. However, for studying cases such as the defibrillation in which the stimuli are applied extracellularly, the bidomain model is the desirable method.
Graphics processing unit (GPU)-based parallelization efforts to this date have been shown to be more effective than parallelization efforts on the CPU-based clusters in terms of addressing the 3D cardiac simulation time challenge. In this project, we investigate both the cardiac cell models and cardiac tissue models in 3D space. We propose algorithmic optimizations based on red black successive over relaxation method for reducing the number of simulation iterations and convergence method for dependence elimination between neighboring cells of the heart tissue. We investigate data transfer reduction and 2D mesh partitioning strategies, evaluate their impact on thread utilization, and propose a strongly scalable cardiac simulation. Our implementation results with reducing the execution time by a factor of five compared to the state-of-the-art baseline implementation. More importantly our implementation is an important step towards achieving real-time cardiac simulations as it achieves the strongest scalability among all other cluster-based implementations.
Publications:
- Ehsan Esmaili, Ali Akoglu, Salim Hariri, “Implementation of Scalable Bidomain Based 3D Cardiac Simulations on a Graphics Processing Unit Cluster,” Journal of Supercomputing, pp. 1-32, March 2019. DOI doi.org/10.1007/s11227-019-02796-8
- Jeno Szep, Ali Akoglu, Salim Hariri, Talal Moukabary, “Two-level Autonomous Optimizations Based on ML for Cardiac FEM Simulations,” 15th IEEE International Conference on Autonomic Computing (ICAC), Trento, Italy, September 3-7, 2018, pp. 101-110.
- Ehsan Esmaili, Ali Akoglu, Gregory Ditzler, Salim Hariri, Jeno Szep and Talal Moukabary, “Autonomic Management of 3D Cardiac Simulations,” IEEE International Conference on Cloud and Autonomic Computing (ICCAC), Tucson, USA, September 18-22, 2017, pp.1-9. (Best Paper)
- Venkata
Krishna Nimmagadda, Ali Akoglu, Salim Hariri, and Talal Moukabary,
"Cardiac simulation on multi-GPU platform," Journal
of Supercomputing, vol 59, no. 3, pp. 1360-1378, 2011. http://dx.doi.org/10.1007/s11227-010-0540-x
- Yoon
Kah Leow, Ali Akoglu, Ibrahim Guven and Erdogan Madenci, "High
performance linear equation solver using NVIDIA graphical processing
units," IEEE NASA/ESA Conference on Adaptive Hardware and
Systems (AHS), San Diego, CA, Jun. 6-9, 2011, pp. 367-374.
C)
High-Speed, Reconfigurable SIGINT (HiReS) System for Large Time- Bandwidth Product
Funded by: ONR
Collaborator: EpiSys Science
Student: Nilangshu Bidyanta, Scott Mashall, Joshua Mack
Software defined radio (SDR) is a technology that has allowed radios, whose functionality has historically been pre-determined by its containing hardware, be defined using software. This technology, which grants the ability for a radio to rapidly reconfigure itself to optimize performance based on the scenario and objectives, has largely enabled the vision of cognitive radio. These radios contain embedded intelligent agents for sensing the environment. An example role of this intelligence is its ability to allow radios share wireless resources without causing interference through sensing the presence of another radio transmission. As this technology has evolved, the ability for these SDRs to analyze their environment has also increased. Instead of simply discerning whether or not a transmission is present, SDRs are beginning to discern between different types of signals by classifying the modulation, or the manner in which information is being relayed. This task of determining the modulation scheme has been given the name modulation classification (MC), recognition, or identification and has a variety of applications in both the military and commercial domains. For example, modulation classification has been proposed as a solution for meeting higher spectral efficiency requirements for the 5G communication systems, dynamic spectrum access networks, and intelligent power control.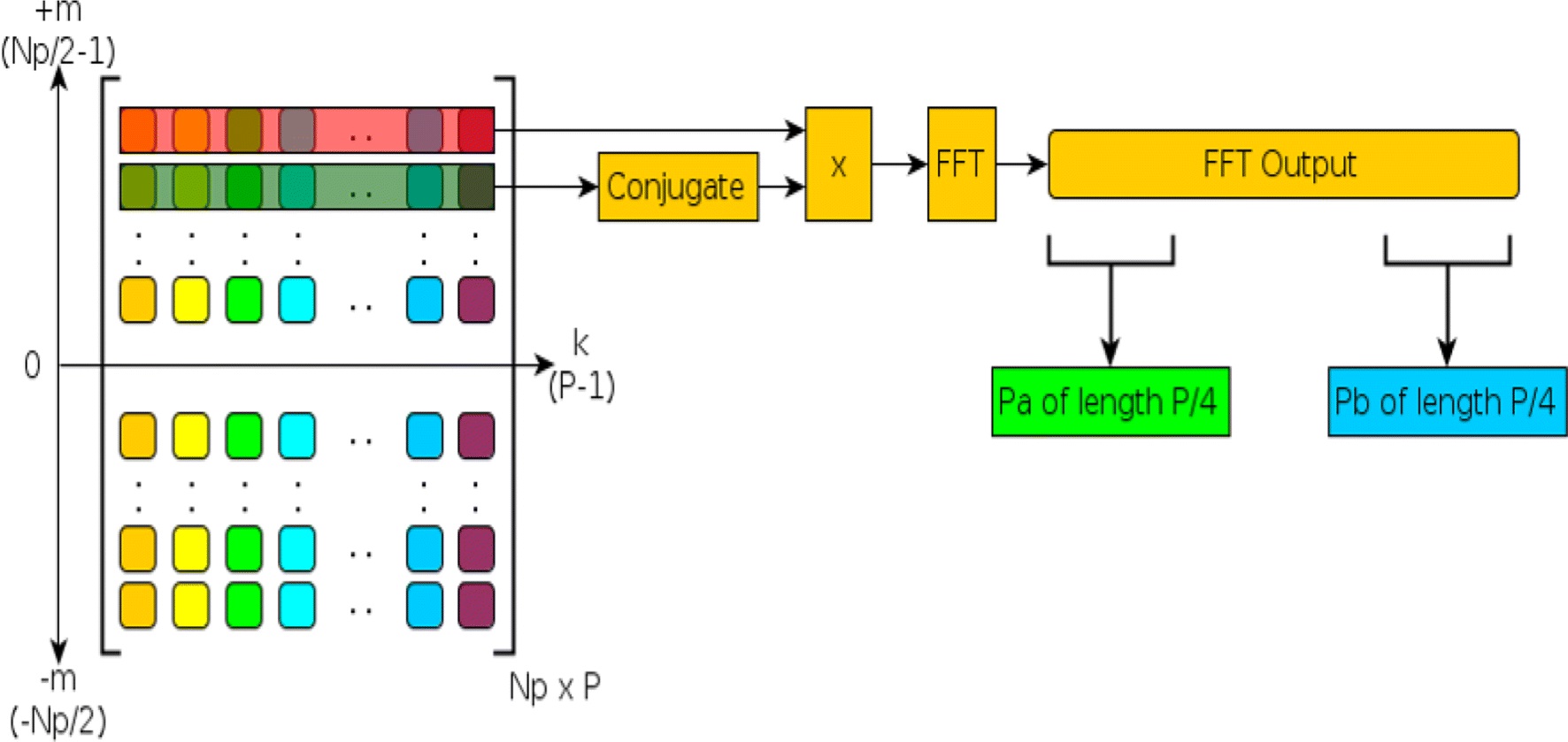 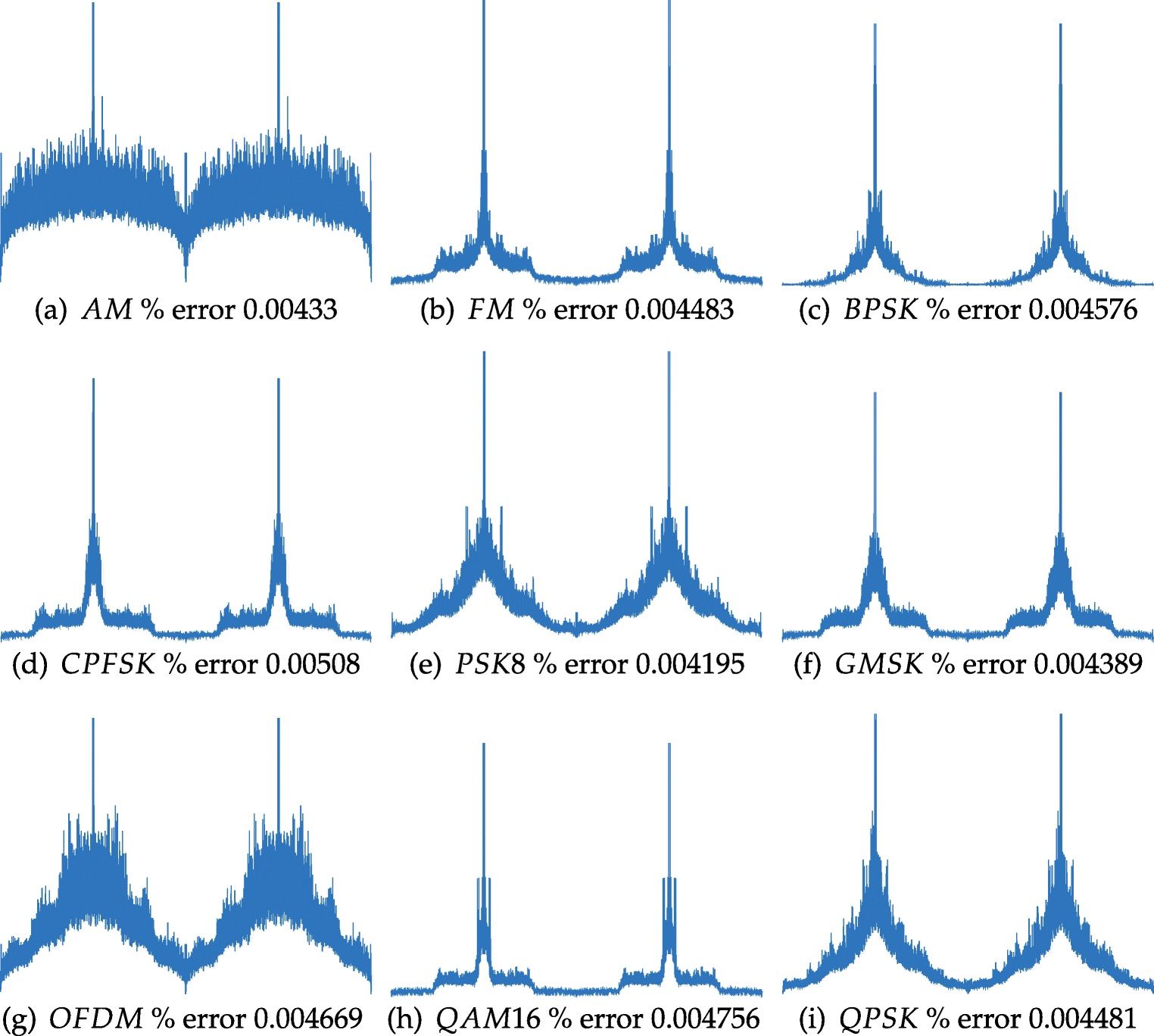
In a previous study where the SCD was parallelized using a GPGPU by primarily exploiting the loop-level data parallel computations in the most compute intensive stages of the execution flow, a throughput of 111 signals per second on Nvidia K20x GPU and a speedup of 390x over the serial version was achieved. We refer to this earlier work as the Baseline SCD implementation [1].
Later we explored a series of algorithmic and architecture-specific optimizations and incrementally quantify the impact of these optimization strategies on throughput. In the follow up study, we show that the kernel launch overhead directly affects the computational throughput and is linked to the sub-optimal thread utilization of the GPU by memory latency and the ratio of time spent on computation in a kernel to the amount of data transferred for kernel instructions and signal data. To overcome this obstacle, we proposed a solution of signal batching by combining the data of many signals into a single kernel call. This allowed for an increase in the number of computations per kernel, resulting in a decrease of overhead associated with launching the kernel. The benefits of this approach are twofold. First, signal batching not only reduces the kernel launch overhead, but also allows for a greater level of data parallelism by concurrently executing multiple signals during each iteration of the SCD flow as opposed to one signal at a time flow of the baseline implementation. Batching queues more work on the GPU for computation, exploiting better overall resource utilization exhibiting an overall speedup of 4.2x using this approach alone compared with the Baseline SCD implementation. Since batching enabled implementation increases the data level parallelism, the potential of utilizing CUDA streams for achieving a more effective distribution of workload is considered as kernels of the SCD flow require varying amount of thread blocks. We present the optimization approach through detailed experimental trend analysis on throughput performance with respect to the changes in batch size and stream size using the Nvidia Tesla K40 GPU. We show that when combined with the signal batching approach, streaming allows an additional performance boost resulting in a combined total speedup of 10.7x by reaching feature extraction capacity for 1282 signals per second. This improved implementation that includes signal batching is referred to as the Full SCD [2].
We further optimized Full SCD implementation by exploiting the symmetry in the SCD that is often used for classification purposes [3]. To exploit this symmetry, a data trimming concept that involves processing a quarter of the input signal data during the iterative FFT stage of the SCD flow, which constitutes 86% of the total execution time, is introduced. The data trimming frees a large amount of thread blocks on the GPU, and hence allows for those threads to be utilized for even more signals. Instead of processing one frame of a given signal at a time, multiple frames can be processed concurrently using multi stream asynchronous execution. We refer to this further-improved implementation as the QSCD, which stands for quarter SCD. Experimental studies are conducted on the QSCD to identify the optimal batch and stream size configuration for effectively utilizing the GPU resources. We show that batching and streaming enabled QSCD implementation improves the throughput from 1282 signals per second to 2719 signals per second on the Nvidia K40 GPU. We also experimentally show that at peak performance, the Full SCD implementation delivers 1569 signals per second, while QSCDdelivers 3300 signals per second and a combined total speedup of 27.5x over the Baseline SCD.
Publications:
- [1] Nilangshu Bidyanta, Garrett Vanhoy, Mohammed Hirzallah, Ali Akoglu, and Bo Ryu, “GPU and FPGA Based Architecture Design for Real-time Signal Classification,” In Proceedings of the 2015 Wireless Innovation Forum Conference on Wireless Communications Technologies and Software Defined Radio (WInnComm’15), March 24-26, 2015, San Diego, CA, pp. 70-79
- [2] Scott Marshall, Garrett Vanhoy, Ali Akoglu, Tamal Bose and Bo Ryu, “GPU based Quarter Spectral Correlation Density Function,” Conference on Design and Architectures for Signal and Image Processing (DASIP), Porto, Portugal, October 10-12, 2018, pp. 88-93
- [3] Scott Marshall, Garrett Vanhoy, Ali Akoglu, Tamal Bose and Bo Ryu, “GPGPU based Parallel Implementation of Spectral Correlation Density Function,” Journal of Signal Processing Systems, pp.1-23, April 2019. DOI doi.org/10.1007/s11265-019-01448-7
Archived projects can be found here and here |




